Chapter 4 Logistic Regression (Binary)
Sources for this chapter:
- R for Marketing Research ad Analytics, Second Edition (2019). Chris Chapman and Elea McDonnell Feit
- ggplot2: https://ggplot2.tidyverse.org/
Data for this chapter:
The
directmktgdata is used from theMKT4320BGSUcourse package. Load the package and use thedata()function to load the data.
4.1 Introduction
Base R is typically sufficient for performing the basics of logisitic regression, but to get some of the outputs required, I have created some user defined functions that are available in the course package.
or_table.R()produces Odds Ratio Coefficientslogreg_cm()produces the Classification Matrixlogreg_roc()produces the ROC Curvesgainlift()produces Gain/Lift tables and chartslogreg_cut()produces the Sensitivity/Specificity Plot
4.2 The glm() Function
glmstands for Generalized Linear Model, and this function can be used with a variety of dependent variables by specifying differentfamily="FAMILY"options *lm(dv ~ iv1 + iv2, data)is the same as
`glm(dv ~ iv1 + iv2, data, family=“gaussian”)- Usage:
glm(formula, data, family)
4.3 Logistic Regression using glm()
Binary logistic regression is performed using the
glm()function when thefamily="binomial"is specified.Usage:
glm(formula, data, family="binonmial")family="binomial"tells R to use logistic regression on a binary dependent variable
If
glm(formula, data, family="binomial")is run by itself, R only outputs the **Logit formulation* coefficients (and some other measures)Call: glm(formula = buy ~ age + gender, family = "binomial", data = directmktg) Coefficients: (Intercept) age genderFemale -8.02069 0.18954 -0.09468 Degrees of Freedom: 399 Total (i.e. Null); 397 Residual Null Deviance: 521.6 Residual Deviance: 336.1 AIC: 342.1Table 4.1: Logit Coefficients from
glm()call
Video Tutorial: glm() Basic Usage
However, if the results of the
glm()call are assigned to an object, the Base Rsummary()function can be used to get much more detailed output, but requires manual calculation of McFadden’s Pseudo-\(R^2\)Call: glm(formula = buy ~ age + gender, family = "binomial", data = directmktg) Coefficients: Estimate Std. Error z value Pr(>|z|) (Intercept) -8.02069 0.78700 -10.191 <2e-16 *** age 0.18954 0.01926 9.842 <2e-16 *** genderFemale -0.09468 0.27354 -0.346 0.729 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 (Dispersion parameter for binomial family taken to be 1) Null deviance: 521.57 on 399 degrees of freedom Residual deviance: 336.14 on 397 degrees of freedom AIC: 342.14 Number of Fisher Scoring iterations: 5# Manually calculate McFadden's Pseudo R-sq Mrsq <- 1-prelim$deviance/prelim$null.deviance cat("McFadden's Pseudo-Rsquared = ", Mrsq)McFadden's Pseudo-Rsquared = 0.3555239Table 4.2: Summary results from
glm()call
For “nicer” looking results, the package
jtoolscan be usedlibrary(jtools) summ(prelim, # Saved object from before digits=4, # How many digits to display in each column model.info = FALSE) # Suppress extraneous informationχ²(2)
185.4317
Pseudo-R² (Cragg-Uhler)
0.5092
Pseudo-R² (McFadden)
0.3555
AIC
342.1413
BIC
354.1157
Est.
S.E.
z val.
p
(Intercept)
-8.0207
0.7870
-10.1915
0.0000
age
0.1895
0.0193
9.8422
0.0000
genderFemale
-0.0947
0.2735
-0.3461
0.7293
Standard errors: MLE
Table 4.3: Summary results from
glm()usingjtoolspackage
Video Tutorial: glm() Output Using the jtools Package
4.3.1 Odds Ratio Coefficients
To get the Odds Ratio coefficients, use the
or_table()function from theMKT4320BGSUcourse package.Usage:
or_table(MODEL, DIGITS, CONF)MODELis the name of the object with results of theglm()callDIGITSis the number of digits to round the values to in the tableCONFis the confidence interval level (e.g., 95 = 95%)
or_table(prelim, # Saved logistic regression model from above 4, # Number of digits to round output to 95) # Level of confidenceParameter OR Est p 2.5% 97.5% (Intercept) (Intercept) 0.0003 0.0000 0.0001 0.0015 age age 1.2087 0.0000 1.1639 1.2552 genderFemale genderFemale 0.9097 0.7293 0.5322 1.5550Table 4.4: Odds Ratio Coefficients
Odds Ratio coefficients can also be obtained from the
summfunction of thejtoolspackagesumm(prelim, # Saved object from before digits=4, # How many digits to display in each column exp = TRUE, # Obtain exponentiated coefficients (i.e., Odds Ratio) model.info = FALSE) # Suppress extraneous informationχ²(2)
185.4317
Pseudo-R² (Cragg-Uhler)
0.5092
Pseudo-R² (McFadden)
0.3555
AIC
342.1413
BIC
354.1157
exp(Est.)
2.5%
97.5%
z val.
p
(Intercept)
0.0003
0.0001
0.0015
-10.1915
0.0000
age
1.2087
1.1639
1.2552
9.8422
0.0000
genderFemale
0.9097
0.5322
1.5550
-0.3461
0.7293
Standard errors: MLE
Table 4.5: Odds Ratio Coefficients using
jtoolspackage
4.4 Estimate with Training Sample Only
Generally, it is good practice to use a training sample and a testing/holdout sample to see how well the model performs “out of sample”. To do this, the data must be split into two groups: train and test
4.4.1 Creating train and test Samples
- While this can be done in a number of ways, the package
carethas a very useful function that attempts to balance the percent of “positive” cases in both samples, while still creating random samples - After running this procedure, two new data frames will be created
- One containing the training data,
train - One containing the testing/holdout data,
test
- One containing the training data,
- Two methods are available: using the
splitsamplefunction from theMKT4320BGSUpackage or manually usingcaret- To use the
splitsamplefunction- Usage:
splitsample(data, outcome, p=0.75, seed=4320)datais the name of the dataframe to be used to estimate the modeloutcomeis the name of the outcome variable (i.e., dependent variable) for the model to be estimatedpis the percent of the data to be assigned to the training sample; defaults to 0.75seedis an integer to be used for random number generation; defaults to 4320
- Requires the
caretpackage - Will create two new dataframes in your environment:
trainandtest
- Usage:
- To use the
caretpackage manually, follow the code below
# Use 'caret' package to create training and test/holdout samples # This will create two separate dataframes: train and test library(caret) # Load the 'caret package' # Start by setting a random number seed so that the same samples # will be created each time set.seed(4320) # Create a dataframe of row numbers that should be in the 'train' sample inTrain <- createDataPartition(y=directmktg$buy, # Outcome variable p=.75, # Percent of sample to be in 'train' list=FALSE) # Put result in matrix # Select only rows from data that are in 'inTrain'; assign to 'train' train <- directmktg[inTrain,] # Select only rows from data that not in 'inTrain'; assign to 'test' test <- directmktg[-inTrain,] - To use the
4.4.2 Run Model with only Training Sample
- Once completed, run the model on the training sample only
- Goodness-of-Fit measures can be obtained from the results
model <- glm(buy ~ age + gender, train, family="binomial") summ(model, # 'jtools' results; use 'summary' for Base R 4, model.info=FALSE)χ²(2) 130.57 p 0.00 Pseudo-R² (Cragg-Uhler) 0.48 Pseudo-R² (McFadden) 0.33 AIC 268.37 BIC 279.49 Est. S.E. z val. p (Intercept) -7.71 0.87 -8.81 0.00 age 0.18 0.02 8.52 0.00 genderFemale -0.01 0.31 -0.04 0.97 Standard errors: MLE; Continuous predictors are mean-centered and scaled by 1 s.d. The outcome variable remains in its original units. Parameter OR Est p 2.5% 97.5% (Intercept) (Intercept) 0.0004 0.000 0.0001 0.0025 age age 1.1996 0.000 1.1505 1.2509 genderFemale genderFemale 0.9873 0.967 0.5399 1.8056Table 4.6: Logistic Regression Results for Training Sample
4.5 Classification Matrix
To get the Classification Matrix, use the
logreg_cm()function from theMKT4320BGSUcourse package.Usage:
logreg_cm(MOD, DATA, "POS", CUTOFF=)MODis the name of the object with results of theglm()callDATAis the data set for which the Classification Matrix should be produced (i.e., original, training, or testing)"POS"is the level of the factor variable that is the SUCCESS levelCUTOFF=is the cutoff threshold; default is 0.5
This function requires the package
caret, which should already be loaded from the splitting of the data# Classification Matrix for training sample logreg_cm(model, # Name of the results object train, # Use training sample "Yes") # Level of 'buy' that represents success/trueConfusion Matrix and Statistics Reference Prediction No Yes No 179 37 Yes 14 71 Accuracy : 0.8306 95% CI : (0.7833, 0.8712) No Information Rate : 0.6412 P-Value [Acc > NIR] : 3.179e-13 Kappa : 0.6136 Mcnemar's Test P-Value : 0.002066 Sensitivity : 0.6574 Specificity : 0.9275 Pos Pred Value : 0.8353 Neg Pred Value : 0.8287 Prevalence : 0.3588 Detection Rate : 0.2359 Detection Prevalence : 0.2824 Balanced Accuracy : 0.7924 'Positive' Class : Yes PCC = 53.99%Table 4.7: Classification Matrix for Training Sample
4.6 ROC Curve
To get the ROC curve, use the
user defined function ``logreg_roc()function from theMKT4320BGSUcourse package.Usage:
logreg_roc(MOD, DATA)MODis the name of the object with results of theglm()callDATAis the data set for which the ROC should be produced (i.e., original, training, or testing)
This function requires the package
pROC, which needs to be loaded# Load 'pROC' package library(pROC) # ROC Curve for the training sample logreg_roc(model, # Name of the results object train) # Use training sample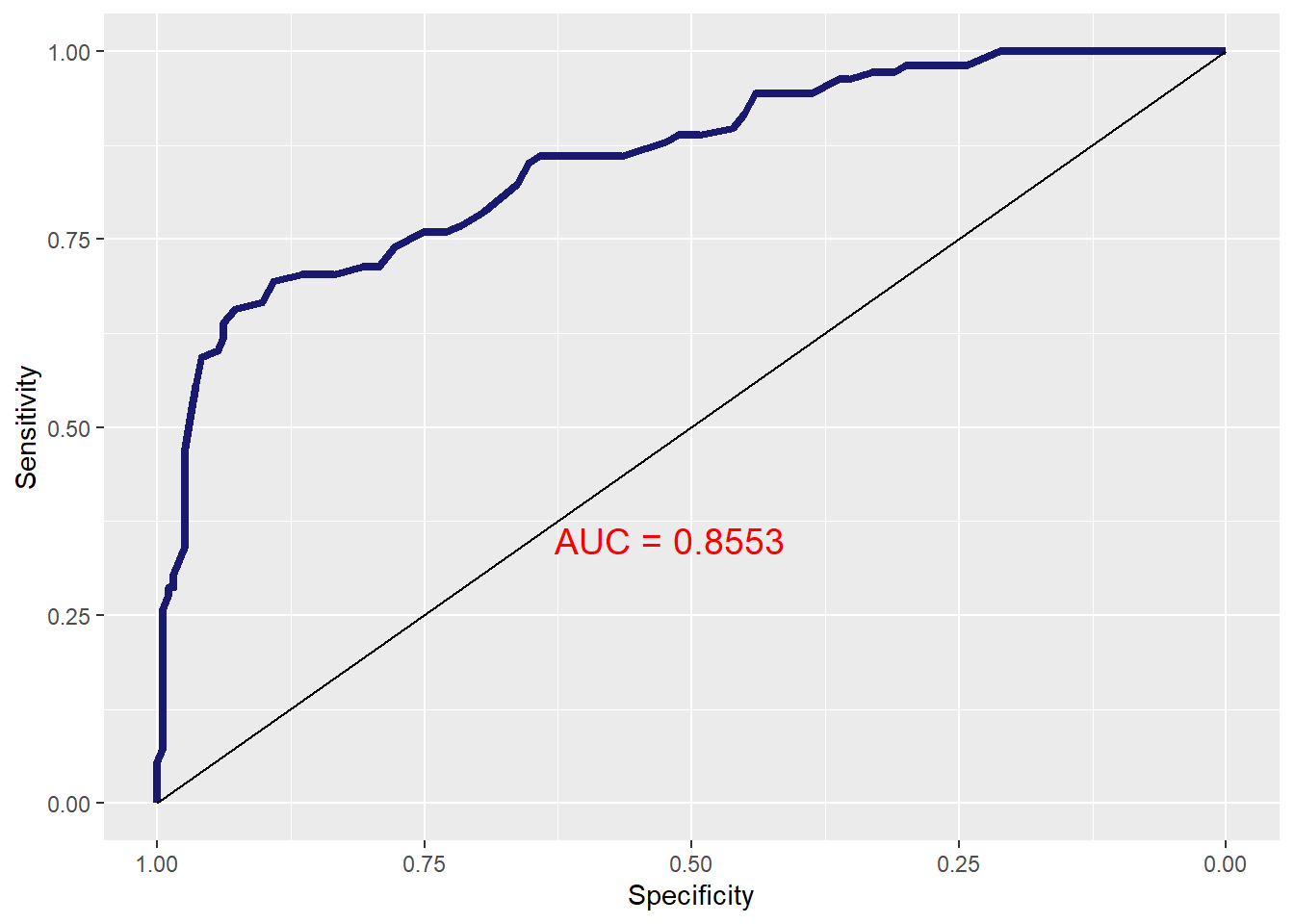
Figure 4.1: ROC Curve for Training Sample
4.7 Gain/Lift Charts and Tables
To get the Gain/Lift charts and tables , use the
gainlift()function from theMKT4320BGSUcourse package.Usage:
OBJ.NAME <- gainlift(MOD, TRAIN, TEST, "POS")OBJ.NAMEis the name of the object to assign the results toMODis the name of the object with results of theglm()callTRAINis the name of the data frame containing the training sampleTESTis the name of the data frame containing the test/holdout sample"POS"is the level of the factor variable that is the SUCCESS level
This function requires the
ggplot2,dplyr, andtidyrpackages, which need to be loaded firstThis user defined function returns a list of four objects
- To use, assign the result to an object name, and then call one of the four objects returned from the function
# Load necessary packages (if not already loaded) library(ggplot2) library(dplyr) library(tidyr) # Call the function and assign to object named 'glresults' glresults <- gainlift(model, # Name of the glm results object train, # Name of the training data frame test, # Name of the testing data frame "Yes") # Level that represents success/trueOBJ.NAME$gaintablereturns the Gain table
# A tibble: 20 × 3 `% Sample` Holdout Training <dbl> <dbl> <dbl> 1 0.05 0.114 0.130 2 0.1 0.229 0.269 3 0.15 0.371 0.370 4 0.2 0.457 0.5 5 0.25 0.6 0.602 6 0.3 0.714 0.667 7 0.35 0.8 0.704 8 0.4 0.8 0.731 9 0.45 0.829 0.769 10 0.5 0.886 0.815 11 0.55 0.914 0.861 12 0.6 0.943 0.870 13 0.65 0.971 0.889 14 0.7 1 0.944 15 0.75 1 0.954 16 0.8 1 0.972 17 0.85 1 0.991 18 0.9 1 1 19 0.95 1 1 20 1 1 1Table 4.8: Gain Table
OBJ.NAME$gainplotreturns the Gain plot
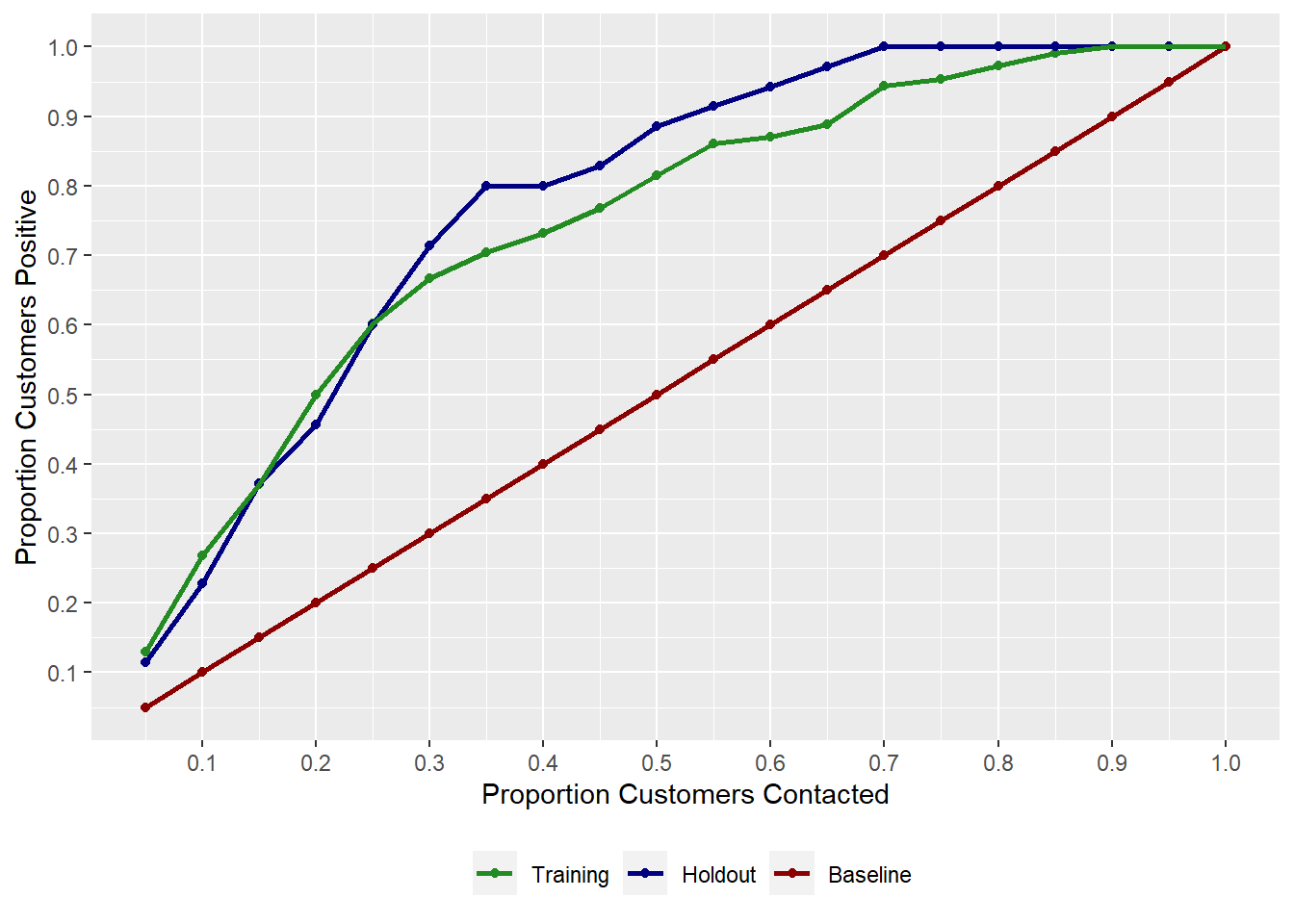
Figure 4.2: Gain Plot
OBJ.NAME$lifttablereturns the Lift table
# A tibble: 20 × 3 `% Sample` Holdout Training <dbl> <dbl> <dbl> 1 0.05 2.83 2.60 2 0.1 2.51 2.69 3 0.15 2.63 2.48 4 0.2 2.38 2.51 5 0.25 2.48 2.42 6 0.3 2.44 2.23 7 0.35 2.33 2.02 8 0.4 2.03 1.83 9 0.45 1.86 1.71 10 0.5 1.79 1.64 11 0.55 1.68 1.57 12 0.6 1.58 1.46 13 0.65 1.50 1.37 14 0.7 1.43 1.35 15 0.75 1.34 1.28 16 0.8 1.25 1.22 17 0.85 1.18 1.17 18 0.9 1.11 1.11 19 0.95 1.05 1.06 20 1 1 1Table 4.9: Lift Table
OBJ.NAME$liftplotreturns the Lift plot
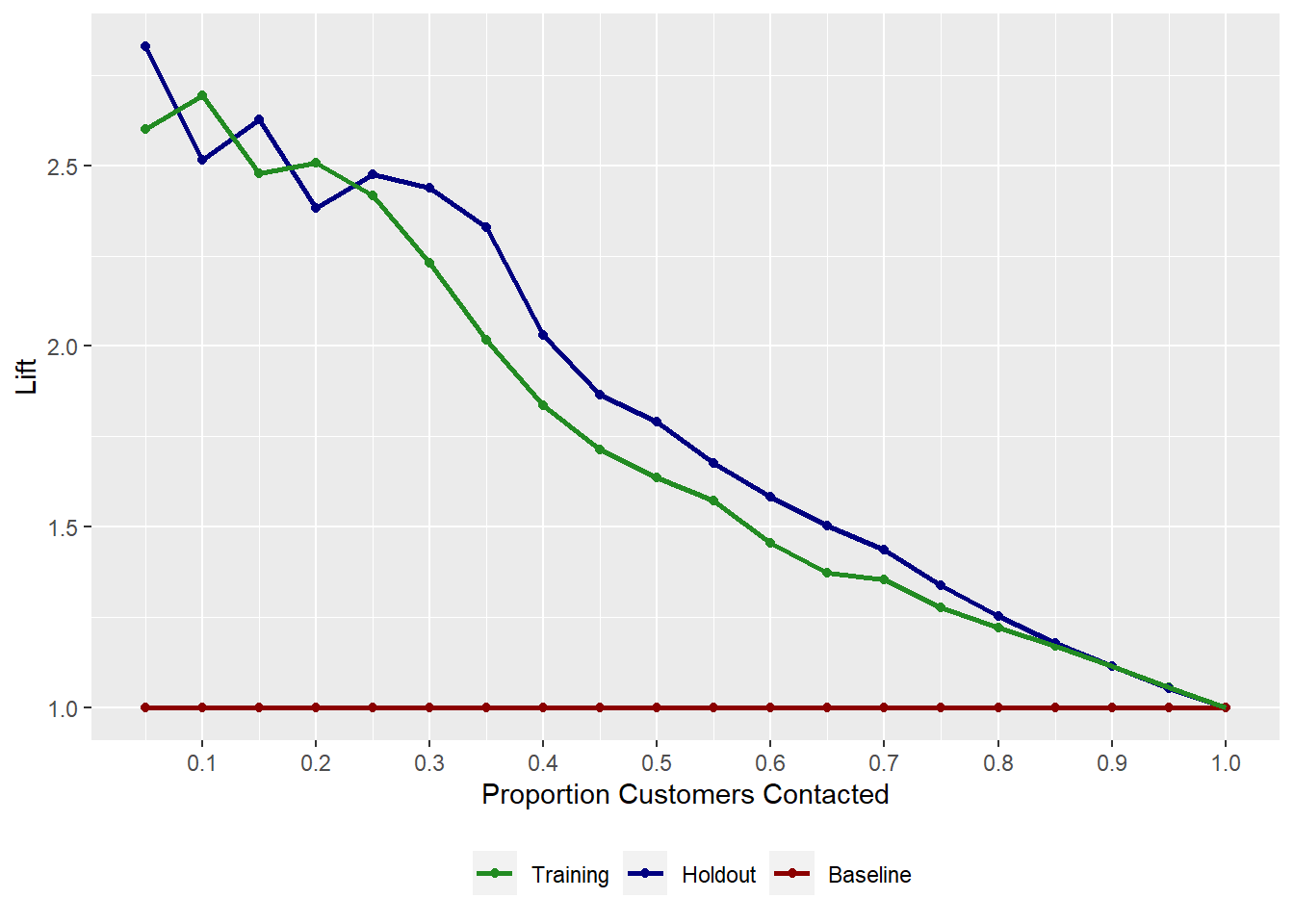
Figure 4.3: Lift Plot
4.8 Sensitivity/Specificity Plot
To get the Sensitivity/Specificity plot, use the
logreg_cut()function from theMKT4320BGSUcourse package.Usage:
logreg_cut(MOD, DATA, "POS")MODis the name of the object with results of theglm()callDATAis the data set for which the plot should be produced (i.e., original, training, or testing)"POS"is the level of the factor variable that is the SUCCESS level
This function requires the
ggplot2package, which needs to be loaded first# Load necessary packages library(ggplot2) # Sensitivity/Specificity Plot for Training Sample logreg_cut(model, # Name of the results object train, # Use training sample "Yes") # Level of 'buy' that represents success/true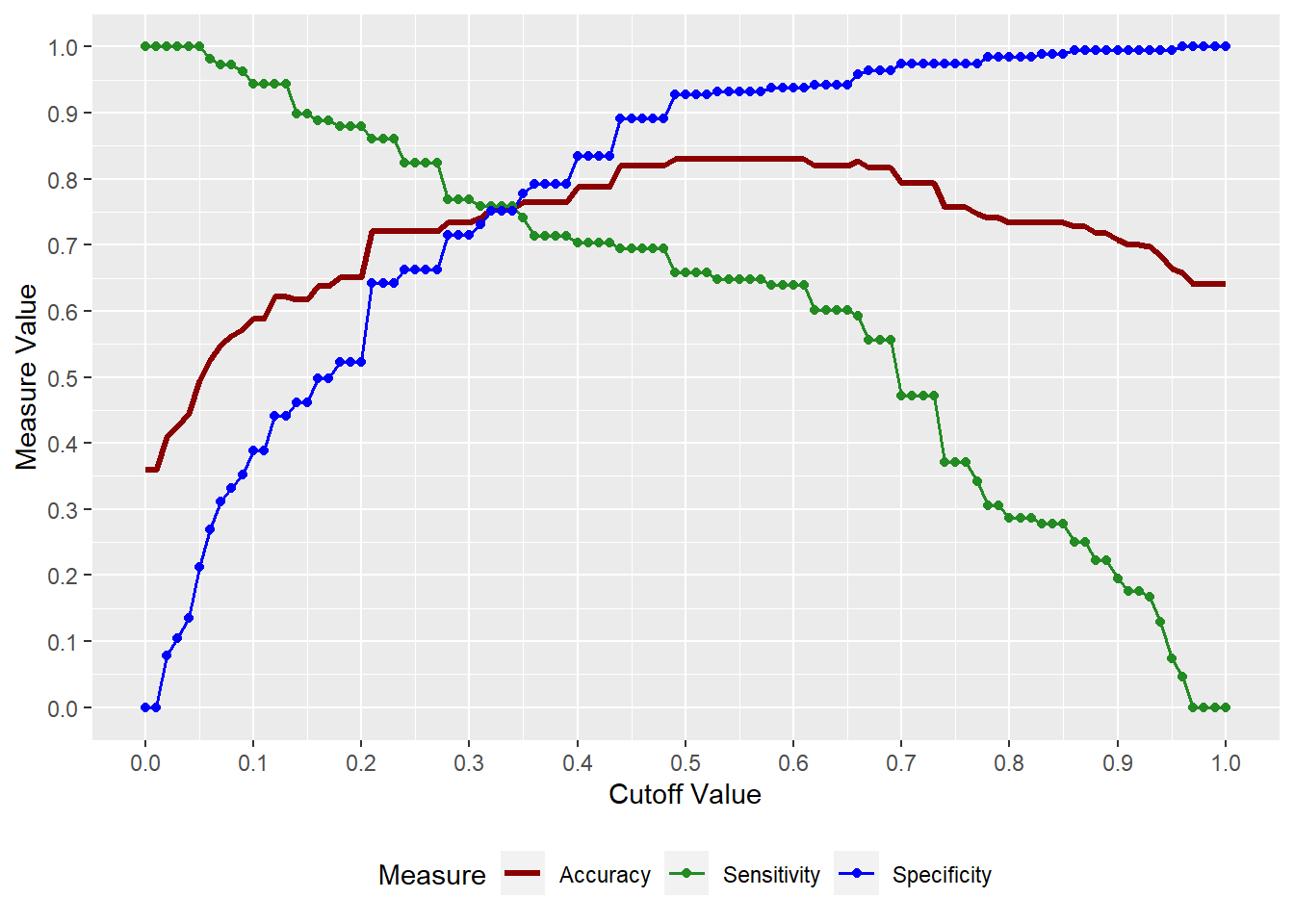
Figure 4.4: Sensitivity/Specificity Plot for Training Sample
Video Tutorial: Logistic Regression - Sensitivity/Specificity Plots
4.9 Margin Plots
Margin plots are done in much the same way as with linear regression margin plots
# Create plot for 'age' and assign to 'p1' p1 <- mymp(model, "age")$plot # Create plot for 'gender' and assign to 'p2' p2 <- mymp(model, "gender")$plot #Use 'cowplot' to put into single plot cowplot::plot_grid(p1,p2)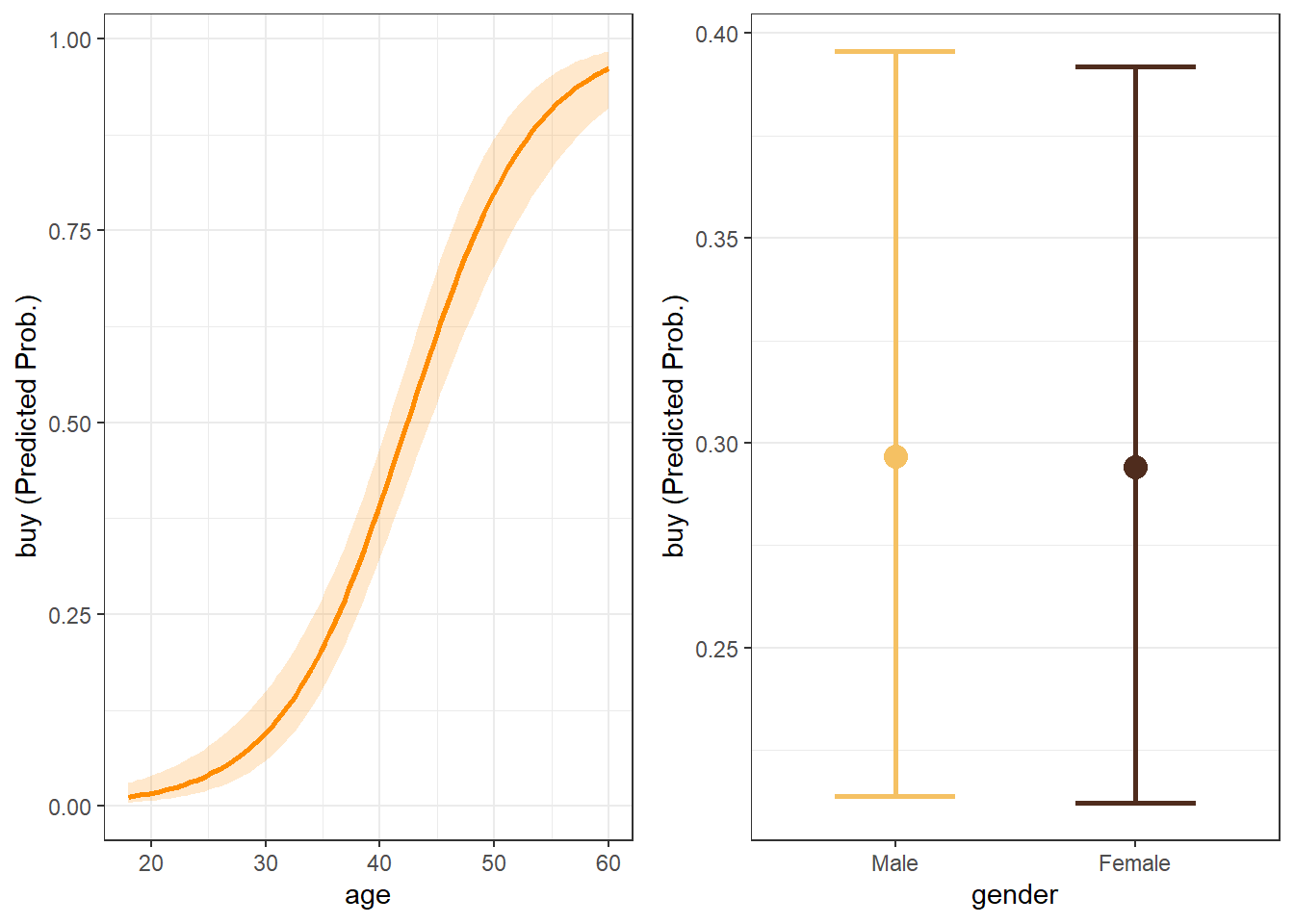
Figure 4.5: Margin plots for variables in model